Module1:Efficacy Verification of AMP
In order to achieve a highly effective and specific targeting of Staphylococcus aureus in the intestine. We have designed an antimicrobial peptide (AMP) - HHHHHHLLGSLLKLLPKLLCASYFCRWWWLL, which has the characteristics of small molecule, targeting and high efficiency in theory. In order to verify its practical effect, we decided to carry out experimental exploration step by step from shallow to deep from chemical synthesis to biosynthesis.
Note: (The pH of the following experimental medium is 8.3).
Iteration1: Chemosynthetic AMP Test
Design
1.Because of the inclusion of modified structures such as targeting domains and His-Tag in the design, we first need to determine whether the designed and synthesized AMP can exert efficient killing efficacy2.Because MRSA is not the only microflora in the gut, we need to ensure that our AMP can kill M RSA in low and efficient quantitieswithout affecting other intestinal colonies too much.
3.Although digestive enzymes are not secreted in the large intestine, residual digestive enzymes are still present in the intestinal fluid. Although we have theoretically demonstrated that there is no digestive enzyme-related site on AMP, further experimental verification is still needed.
Build
1. AMP was chemically synthesized, and an appropriate amount of MRSA was cultured under the condition of 1um concentration of AMP and the same concentration of normal saline, and its growth was observed, and electron microscopy observation and flow cytometry analysis were performed.2. AMP was chemically synthesized, and MRSA, BS, EC, and PA were co-cultured at a concentration of 1um AMP and the same concentration of normal saline, respectively.
3. AMP was chemically synthesized, an appropriate amount of trypsin and intestinal peptidase were added to the medium, and then an appropriate amount of MRSA was cultured at a concentration of 1um AMP and the same concentration of normal saline
Test
The growth of MRSA and other strains after treatment was observed, and electron microscopy and flow cytometry analysis were performed.Iteration2: Biosynthetic AMP Test
Design
After verifying the efficacy of AMP itself, we will perform simulated experiments on biosynthesis in combination with specific application environments and chassis organisms.1.First, we need to verify that chassis organisms can successfully express AMP after modification, and how much AMP can be secreted. Because the amount of AMP secretion is directly related to the effectiveness of our program and the size of side effects.
2.We need to verify that biosynthesized AMP has the same good biological effects as chemically synthesized AMP
3.We are to see if the digestive enzymes and PH in the gut interact with chassis organisms that cause other effects on the object.
Build
1. After the constructed plasmid is introduced into the chassis organism, it is cultured in vitro for a period of time before being purified by immobilized metal ion affinity chromatography (IMAC).2. After the constructed plasmid was introduced into our chassis, the engineered chassis organisms and the non-engineered chassis organisms were co-cultured with MRSA, respectively
3. After the constructed plasmid was introduced into our chassis, an appropriate amount of trypsin and enteropeptidase was added to the culture medium, and then the modified chassis organisms and the non-modified chassis organisms were co-cultured with MRSA
Test: Fourth, verify whether the modified chassis organisms can successfully secrete AMP and determine its secretion concentration
1. Purification by immobilized metal ion affinity chromatography (IMAC) was performed, and purification and concentration determination of AMP were performed with the help of His-Tag
2.Its growth was observed, and electron microscopy and flow cytometry analysis were performed.
Learn
1.If the protein is not expressed (because it is too short, etc.), fluorescence in situ hybridization can be used to see if the corresponding mRNA is expressed and whether the corresponding gene is duplicated2.If the protein is expressed but not potent, the pH of the medium can be changed for gradient testing, and if the AMP is inactive due to pH, further residue substitution can be performed for antimicrobial peptides
3.Protein expression, but no potency, if not pH, may be a structural problem with the chassis biological modification (e.g., lack of disulfide bonds) that can be sequenced for purified proteins
4.If the biosynthetic concentration is higher than required, the addition of silicons may be considered.
5.If the synthetic concentration is lower than desired, repeats or an increase in the number of imported plasmids may be considered
In order to verify whether the antimicrobial peptide we designed can meet the application needs of our project, we designed a test to validate it
References
- L. Shang et al., “Hybrid Antimicrobial Peptide Targeting Staphylococcus aureus and Displaying Anti-infective Activity in a Murine Model,” Front. Microbiol., vol. 11, p. 1767, Sep. 2020, doi: 10.3389/fmicb.2020.01767.
- B. Mishra and G. Wang, “Ab Initio Design of Potent Anti-MRSA Peptides Based on Database Filtering Technology,” J. Am. Chem. Soc., vol. 134, no. 30, pp. 12426–12429, Aug. 2012, doi: 10.1021/ja305644e.
Module2:Sensor Switch and Control System
In order to better achieve the goal of targeted anti-Staphylococcus aureus therapy and reduce antibiotic contamination, we believe that it is necessary to design an efficient, accurate, sensitive and safe control system. Therefore, we designed a highly specific Staphylococcus aureus sensor switch and a feedback mechanism to properly control AMP expression, which needs to have positive feedback response and negative feedback modulation.
Iteration1: Separate Components Test
Design
--Sensor switchBut take into account AgrC protein. As membrane surface receptors require accurate membrane surface representation, we try to construct E. coli surfaceslocalization of the fusion protein of peptide and AgrC,and carry out experimental tests on the surface display of the membrane, including:lnK-N-his tag-AgrC,his tag-AgrCTwo experimental groups。[4]
In order to test the performance of the sensing element, we tried the design of the sfGFP fluorophore attached to the downstream of the P3 promoter and constructed the relevant strains, and experimentally plotted the response curve of the fluorescent reporter gene to the concentration and time, so as to explore the detection domain.
Further investigation showed that lambda phages stably entered the lyogenic state through a series of gene interactions and self-overexpression inhibition of Ci protein after infection. This gave us the inspiration to reinvent the feedback loop.
Cro will express and inhibit the PRM promoter with zero input, and after the input, we designed a fast-response expression pathway with positive feedback characteristics by expressing the Ci protein at the same time, and then through the inhibition of the Ci protein on the promoter PR expressing the cro, and the tandem structure of two reversed-phase devices to relieve its own transcriptional repression, and designed two experiments for simple verification
1.We found that there were reports in the literature that the PRM promoter was weak, and we transferred a simple PRM-sfGFP sequence to explore the promoter efficiency.2.Further, we designed an iPTG-induced sfGFP-containing plasmid to verify the design efficiency of our positive feedback pathway.
There are six Ci protein binding sites, OR1, OR2, OL1, OL2, OR3, and OL3, upstream and downstream of the Ci protein sequence, and the Ci protein will prevent the initiation of transcription here, and OR3 is located on the promoter PRM, Ci does not bind at low concentrations,At high concentrations, it will form a DNA circular structure to promote the formation of self-concentration clamping.[6]Based on this, we designed a negative feedback regulatory pathway (PL-Ci) that clamps AMP expression levels-day-PRM-PR sequences were used for the experimental group) and in a system without ORs, OLs lociAs a control, the clamping concentration and system stability were verified.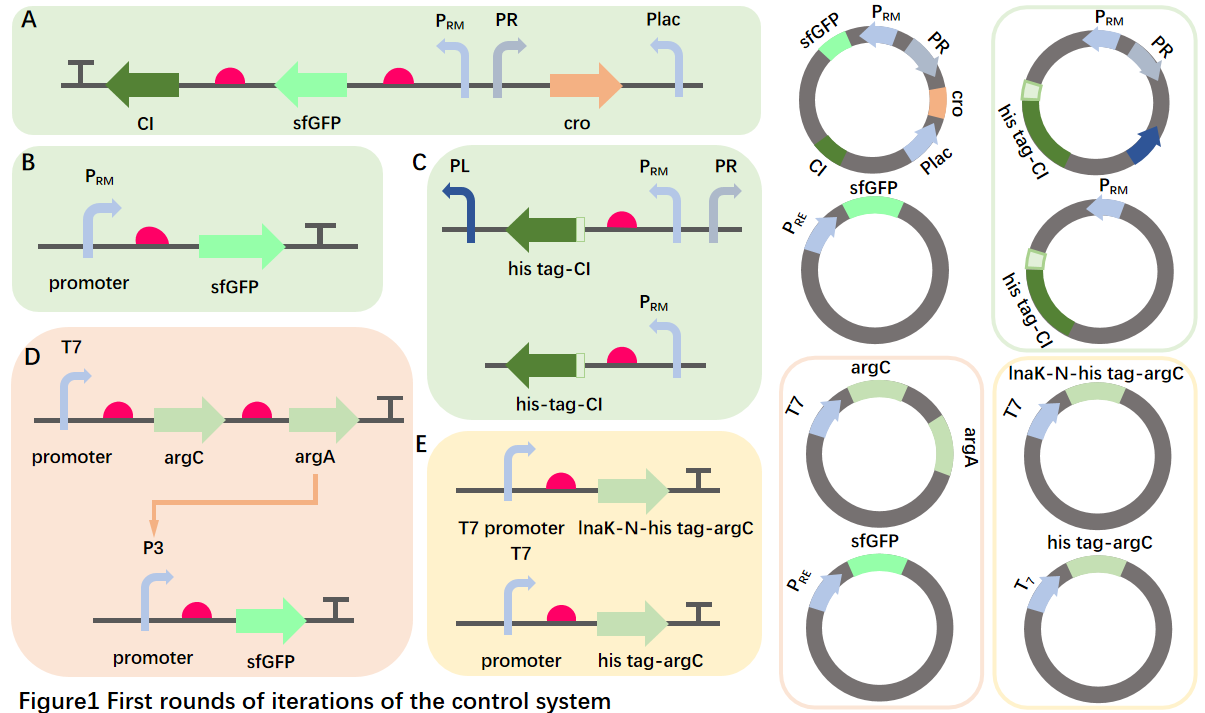
--Sensor switch
First, we will construct and electroporate the transduction plasmid into E. coli to construct the desired strain, and for the two experimental purposes mentioned above:
1.We collect and isolate cells, extract intracellular proteins of different membrane components, prepare samples for western blot analysis, and obtain membrane localization information of the target protein.[4]2.We first cultured in normal medium without AIP to observe the fluorescent protein signal intensity and assess the level of gene leakage, and then introduced a concentration gradient of synthetic AIP and Staphylococcus aureus cell lysates into the culture early in the bacterial culture index and co-cultured for a period of time.
--“EcoTest”
For the liquid bacterial samples to be measured, we first mix the culture medium and take a certain sample, dilute it by a certain factor, measure the absorbance under OD600 of the microplate reader to roughly determine the population density, and take samples at the same time to obtain the gene transcription level by RT-qPCR.
Fluorescence intensity was determined at 488 nm excitation light for samples containing reporter sfGFP, and ELISA method for samples containing his-tag was used to determine concentration.
For a series of concentration gradients or time samples, we used the blank group as the control to calculate the relative fluorescence intensity and plot the protein response expression curve at a certain density, and calculated the relative transcription level and plotted the transcription response curve according to the RT-qPCR results
For specific experimental methods, click the link to go to the protocol
For the response experiment, we calculated the gene leakage expression rate under the 0 concentration A IP group, and furthermore, we used the 0 concentration AIP group as the control, and obtained the concentration response curve with the idea of "EcoTest" to verify the sensitivity and efficiency of the sensor switch
For the validation experiment of the positive feedback pathway, we took the plasmid lacking PR promoter and CRO coding as the control, and after entering the plateau period, iPTG was added to the medium to induce the expression of his tag-CI, and then a batch of samples was taken every 10 minutes.
For the validation experiment of the negative feedback pathway, a batch of samples was taken every 30 minutes after several hours of shaker expansion, and the response curve was obtained by "EcoTest", and it was observed that the experimental group Ci entered the steady state of lower concentration earlier.
--Sensor switch
We have made a preliminary exploration of the detection domain and response curve, and found that the linear/nonlinear response of the following concentration range can be achieved, and the corresponding modeling will be carried out on this basis.
Iteration2:Optimisation of component performance
In the further study of engineering ideas, we realized that in the actual production environment, the system will face many interferences in the complex environment, and we need to consider the robustness of the system and verify and optimize it.
--Sensor switch
At the same time, through the analysis of the negative feedback mechanism, we guessed that changing the length of the OR1-OL1 sequence may affect the final concentration level, and experimentally verified this guess.
--Sensor switch
Anti-crosstalk experiment: We obtained some common gram-positive pathogens in various intestines that grow, such as Staphylococcus aureus, Bacillus subtilis, etc., with Bifidobacterium as a variable, called "sensing bacteria";Firstly, different types of "inductive bacteria" were inoculated in a blank liquid medium, and after the appropriate time was cultivated, each engineering bacteria were inoculated, and the medium without inductive bacteria was used as a control.
Real-time experiments: Literature investigation found that erythromycin and aminobutyric acid can change the ability of bacteria to secrete AIP, and we can add corresponding drugs in the co-culture environment to induce changes in the concentration of A. aureus secretion,so as to observe whether our sensor switch can achieve timely response.
Sequence adjustment experiment: Based on the original plasmid, we designed nonsense nucleotide sequences ranging from 30 bp to 180 bp downstream of Ci, repeated the relevant experiments involved in the first round and verified them by WB.
--Sensor switch
Anti-crosstalk experiment: (assuming good effect) the fluorescence intensity of the other strains was lower than that of the control group with no sensors, while the Staphylococcus aureus group had a significantly stronger fluorescence signal
Real-time experiment: (assuming a good effect) verifies the accuracy of a round of detection domain, in which the fluorescence intensity is sensitively up/down-regulated in a short period of time
Sequence adjustment experiment: The protein expression level and gene transcription level in the steady state were obtained by "EcoTest", and the blot results were analyzed by image J to confirm the protein expression level.
--Sensor switch
If the fluorescence intensity is not accurate, the experiment can be repeated by S DS-PAGE quantification and flow cytometry to determine the fluorescence intensity; it may be the interference of sensing bacteria, and then the bacteria + medium environmental extract can be used as the medium substrate by cultivating the corresponding bacteria, and even synthesizing the A IP sub of different strainstype to carry out the experiment
2.Real-time experiments: It indicates that it has good real-time performance, good targeting, and has the potential to induce Staphylococcus aureus in a complex intestinal microenvironment.
2.1 round: It may be that the expression of Agr protein is not enough, and then the experiment can be repeated by increasing its expression; if the fluorescence intensity detection is not accurate, the experiment can be repeated by S DS-PAGE quantification and flow cytometry to measure the fluorescence intensity; it may be that the experimental quantification method is not accurate enough, the total protein can be extracted by rapid collection and then detected; by RBS, promoter orientation optimization, etc., to obtain sequences with higher expression efficiency.Iteration3: Gene Circuit Assembly
After validating and optimizing each discrete functional element, we try to splice it into a complete gene circuit to build a gene that can autonomously respond to and control exosome AMP engineering bacteria are mainly divided into three steps:
①PRE promoter efficiency verification: Cii, Ciii protein can activate the expression of PRE, which was selected as the signaling molecule that connects the sensing switch to the feedback pathway, and we first constructed the relevant vector and verified the PRE promoter efficiency.②Connection between the sensor switch and the feedback path: Connect the Cii and Ciii to the downstream of the response system to test whether they can be connected.
③Connect the control system to the AMP series: Connect the AMP to the upstream of Ci to test whether the AMP is expressed normally
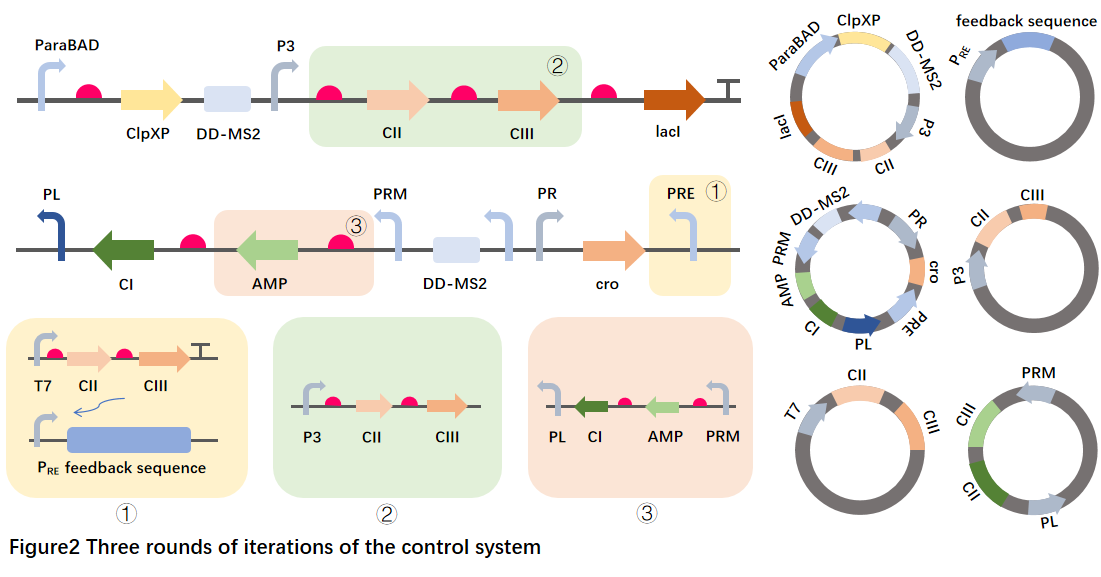
We follow the idea of BioBricks, use the existing plasmid as the backbone to carry out modular connection, step by step to construct the corresponding strains, and repeat the previous experiments under stable culture conditions.
We used the corresponding vector template plasmid as a blank control to determine the relevant response curves to verify whether the overall response and feedback pathway were connected, and whether AMP could be adjusted to the appropriate expression level.
We have successfully constructed an engineered bacteria that release AMP in a controlled manner, and the next step may be to consider more realistic use scenarios, as well as possible problems such as low efficiency and large chassis bioload, we may try to optimize by further directed evolution, streamlining sequences, etc.
Iteration4:Simulation Experiments Combined with Hardware
Considering that the complex factors of application may have an unpredictable impact on the achievement effect, we hope to build an integrated intestinal microarray testing system to test the colonization ability of engineered bacteria, the ability to interact with each other in the intestinal microbiota, and the killing ability of Staphylococcus aureus.
The test system includes an intestinal chip, an electronic measurement system and related processing procedures, and the three modules are directly combined and assembled to build a complete test system.
The specific design details are detailed in our "Hardware" section.
Again, the specific construction details are described in more detail in our "Hardware" section.
A Staphylococcus aureus infection model was constructed by introducing Staphylococcus aureus strains on intestinal microarrays through co-culture.
IVIS imaging was performed at different time points using the fluorescent protein labeling of the engineered bacteria, which showed the distribution and density of E. coli at different stages of infection, and the response of the engineered bacteria to Staphylococcus aureus could be observed. At the same time, at different time points, the latex agglutination test was carried out by taking the outer base culture medium, and the corresponding distribution density of Staphylococcus aureus was obtained, reflecting the killing effect of Escherichia coli on Staphylococcus aureus at the corresponding time points.
The density and distribution of engineered bacteria obtained by hardware can reflect the response effect of E. coli in practical applications, indicating that our design can be verified at the response level of engineered bacteria.
The density of Staphylococcus aureus in the corresponding period can characterize the killing efficiency of Escherichia coli, and verify that the engineered bacteria we designed can have a good killing effect on Staphylococcus aureus in the near in vivo environment, which has a certain degree of reference significance for specific clinical applications.
- Di H., Lan L., and Chen F., “Small molecules targeting Staphylococcus aureus virulence,” Chin. Sci. Bull., vol. 58, no. 36, pp. 3743–3752, Dec. 2013, doi: 10.1360/972013-351.
- D. Lubkowicz, C. L. Ho, I. Y. Hwang, W. S. Yew, Y. S. Lee, and M. W. Chang, “Reprogramming Probiotic Lactobacillus reuteri as a Biosensor for Staphylococcus aureus Derived AIP-I Detection,” ACS Synth. Biol., vol. 7, no. 5, pp. 1229–1237, May 2018, doi: 10.1021/acssynbio.8b00063.
- T. J. Polaske, K. H. J. West, K. Zhao, D. L. Widner, J. T. York, and H. E. Blackwell, “Chemical and Biomolecular Insights into the Staphylococcus aureus Agr Quorum Sensing System: Current Progress and Ongoing Challenges,” Israel Journal of Chemistry, vol. 63, no. 5–6, p. e202200096, Jun. 2023, doi: 10.1002/ijch.202200096.
- S. Bao et al., “Construction of a cell-surface display system based on the N-terminal domain of ice nucleation protein and its application in identification of Mycoplasma adhesion proteins,” J Appl Microbiol, vol. 119, no. 1, pp. 236–244, Jul. 2015, doi: 10.1111/jam.12824.
- J. J. Gierut, T. E. Jacks, and K. M. Haigis, “Strategies to Achieve Conditional Gene Mutation in Mice,” Cold Spring Harb Protoc, vol. 2014, no. 4, p. pdb.top069807, Apr. 2014, doi: 10.1101/pdb.top069807.
- I. B. Dodd, A. J. Perkins, D. Tsemitsidis, and J. B. Egan, “Octamerization of λ CI repressor is needed for effective repression of P RM and efficient switching from lysogeny,” Genes Dev., vol. 15, no. 22, pp. 3013–3022, Nov. 2001, doers: 10.1101/gad.937301.
- S. K. Aoki, G. Lillacci, A. Gupta, A. Baumschlager, D. Schweingruber, and M. Khammash, “A universal biomolecular integral feedback controller for robust perfect adaptation,” Nature, vol. 570, no. 7762, pp. 533–537, Jun. 2019, doi: 10.1038/s41586-019-1321-1.
Module3:Biology Safety
We have designed several layers of biosafety modules based on different levels of life for different safety purposes, and for the above work, we have designed rigorous experiments to explore the biocontainment efficiency of relevant modules.
Iteration1: Separate Components Test
-- DD-SHIELD1 system
The specific design can be found here.
The specific design can be found here.
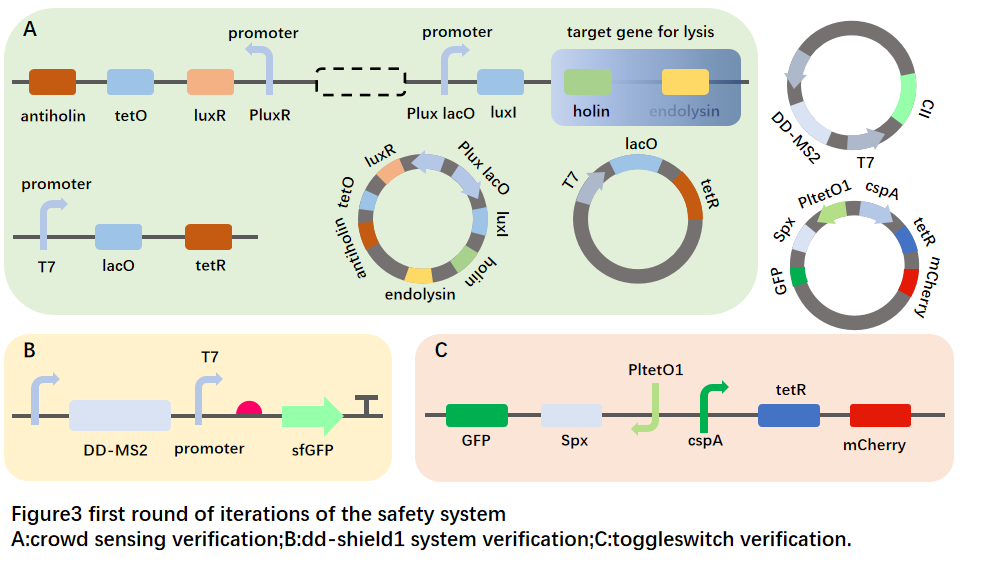
-- DD-SHIELD1 system
On the basis of the above, the strains constructed with toxin proteins entered the plateau stage, centrifuged, collected bacteria, and resuspended at twice the amount of bacteria or twice the amount of medium.
-- DD-SHIELD1 system
Through multi-level biosafety transformation, we have built a good biocompatible engineering chassis, but whether excessive modification will cause inefficiency of functional modules and limited application scenarios. In the future, we will consider further designing safety systems with more controllable performance.
Iteration2:Optimisation of Component Performance
--Quorum sensing system
The updated gene circuit design can be found here.
The specific design can be found here.
--Quorum sensing system
--Quorum sensing system
Luckily, the verification of toggle switch has been successful, which has a strong conversion efficiency, a lower leakage rate, a faster conversion speed, and better meet the effect of engineering bacteria production and taking. For its existing shortcomings, we will consider replacing it with a higher efficiency promoter to improve the conversion efficiency, changing a stronger terminator, to reduce gene leakage and other ways to improve.
Our new quorum sensing system tries to take into account the cellular burden and good functional implementation, and can manually change the set population threshold, which broadens the use scenarios of the system and facilitates the more refined regulation of colony density.
- J. Zhang et al., “Inhibition of quorum sensing serves as an effective strategy to mitigate the risks of human bacterial pathogens in soil,” Journal of Hazardous Materials, vol. 465, p. 133272, Mar. 2024, doi: 10.1016/j.jhazmat.2023.133272.
- C. Ge et al., “Redesigning regulatory components of quorum-sensing system for diverse metabolic control,” Common Nat, vol. 13, no. 1, p. 2182, Apr. 2022, doi: 10.1038/s41467-022-29933-x.
Module 4: Part about Bacterial Hydrogels
It consists of three test cycles for the construction of a hydrogel system that can be secreted by E. coli.
Iteration1 : Measurement of hydrogel biosynthetic amount
Iteration2 : Tests of the physicochemical properties of hydrogels
Iteration3 : Engineered bacteria adhesion ability and safety test
Iteration1
Understanding the key role of the mucosal system outside the intestinal epithelial cells in intestinal homeostasis, we hope to construct a hydrogel that can be secreted by microorganisms to antagonize the colonization of pathogenic bacteria, repair the intestinal mucosal barrier, and enhance the adhesion and colonization ability of engineered bacteria.
To this end, referring to previous studies, we designed a triplet chimeric protein, the frizzling protein CsgA, the tag 6×His for ligation and examination, and the cytokine TFF, based on the trilobite factor (TFF) of the fibrous matrix of the intestinal mucosa itself. It was expected that TFF would be tethered to a crimped fiber matrix to construct a self-crimping hydrogel.
Among them, CsgA itself is a common protein component in the extracellular matrix of bacteria, and is a class of frizzled proteins that can be used as scaffolds for active biomaterials.
TFF, on the other hand, is a small protein secreted by mucus-producing cells on the surface of the gastrointestinal tract and other mucous membranes, which can be used to promote epithelial recovery and inhibit intestinal inflammation. According to the different TFFs, three chimeric proteins, cTFF1, cTFF2 and cTFF3, were designed for subsequent production and testing.
In order to achieve successful modification and secretion of chimeric proteins, they need to be co-transcribed with the necessary modification genes (csgB, csgC, csgE, csgF, csgG). Therefore, we co-composed their genes into a coiled protein expression operon that wasintroduced into the kanamycin select-tagged pBbB8k plasmid backbone using P BAD as the promoter.
The plasmid was electrotransduced into E. coli to obtain the corresponding three types of strains. The transformed bacteria are cultured at 37 °C in high osmolality medium to mimic physiological conditions and induced with arabinose.
Using the quantitative Congo red binding (CR) assay, the expression efficiency of different fusion methods could be investigated to explore whether the three CsgA-TFF fusion proteins could be expressed and assembled into crimped fibers under predetermined conditions under wild-type control.
Using a whole-cell enzyme-linked immunosorbent assay (ELISA), the 6xHIS tag was probed to further confirm the extracellular assembly.
Through this round of experiments, it was confirmed that the chimeric proteins we designed could be expressed and assembled as crimped fibers under predetermined conditions, demonstrating the usability of the hydrogel system.
In addition, the expression and assembly efficiency of chimeric proteins of different designs were preliminarily shown through control experiments, which provided ideas for the selection and optimization of final hydrogel proteins.
Iteration2
Through this round of experiments, it was confirmed that the chimeric proteins we designed could be expressed and assembled as crimped fibers under predetermined conditions, demonstrating the usability of the hydrogel system.
Bacterial cultures are vacuum-filtered to obtain concentrated biomass. After treatment with 5% SDS solution, wash well with water. The concentrated hydrogel is scraped on the membrane and consists of 95–97% water. This hydrogel can be stored in a humid environment at 4°C or lyophilized into powder form to meet our application requirements.
Scanning electron microscopy (SEM) was used to characterize the hydrogels, which could reveal the gelatinization effect and the network structure of the hydrogels at a microscopic level. Rheological frequency sweeps can understand the response of a material to impact or gradual loading. The storage modulus and loss modulus of the hydrogel can be obtained by rheological scanning, and the mechanical properties of the gel such as inertia, impact resistance, and damping properties can be understood.
The SEM results of the samples show the nanostructure of the hydrogel material, which can prove the successful gel formation of the designed hydrogel, and at the same time, the excellent biomimetic properties of the material can be verified by comparing it with the natural structure. The results of rheological analysis can show the excellent physical and chemical properties of hydrogels, and the influence of different factors on the properties of the materials can be probed, which can provide a reference for the adjustment under actual application.
Iteration3
Considering the need for long-lasting colonization of engineered bacteria in specific use cases, it is necessary to evaluate the adhesion characteristics and safety of live bacteria hydrogels. To this end, we tried to simulate different levels of the biological environment, combined with the design of hardware, to study the different properties of hydrogels.
Simulates the construction of the gut, and the specific details are elaborated in more detail in the "Hardware" section.
The adhesion characteristics of the gel, the residence of the gel and the colonization ability of the engineered bacteria could be characterized by labeling the live gel with Cy7, staining with Congo red, visualizing the gel, and imaging the simulated intestine with IVIS at different time points.
Co-culturing engineered bacteria in an in vitro model of a human colorectal adenocarcinoma cell line (Caco-2) can confirm whether overproduction of modified crimped fibers induces a pathogenic phenotype of PBP8.
The results of gel adhesion experiments can prove the effectiveness of the introduction of hydrogel system in enhancing the adhesion ability of engineered bacteria, indicating that hydrogels and engineered bacteria can achieve long-term continuous colonization in the intestine, which verifies the feasibility of the desired long-lasting colonization effect of the intestinal tract to a certain extent.
The co-culture experiments of Caco-2 showed that the loaded engineering bacteria had low translocation and low invasiveness, and the overproduction of frizzled protein did not induce the pathological phenotype, which provided a certain degree of guarantee for the safety of clinical administration.
- Duraj‐Thatte, Anna M, et al. “Hydrogels: Genetically Programmable Self‐Regenerating Bacterial Hydrogels (Adv. Mater. 40/2019).” Advanced Materials, vol. 31, no. 40, 1 Oct. 2019, https://doi.org/10.1002/adma.201970289. Accessed 17 Feb. 2024.
- Duraj‐Thatte, Anna M., et al. “Genetically Programmable Self‐Regenerating Bacterial Hydrogels.” Advanced Materials, vol. 31, no. 40, 12 Aug. 2019, p. 1901826, https://doi.org/10.1002/adma.201901826.
- Hwang, In Young, et al. “Engineered Probiotic Escherichia Coli Can Eliminate and Prevent Pseudomonas Aeruginosa Gut Infection in Animal Models.” Nature Communications, vol. 8, no. 1, Apr. 2017, www.nature.com/articles/ncomms15028, https://doi.org/10.1038/ncomms15028. Accessed 1 Jan. 2020.
- Mays, Zachary J. S., et al. “Quantifying and Engineering Mucus Adhesion of Probiotics.” ACS Synthetic Biology, vol. 9, no. 2, 7 Jan. 2020, pp. 356–367, https://doi.org/10.1021/acssynbio.9b00356. Accessed 6 Mar. 2022.
- Praveschotinunt, Pichet, et al. “Engineered E. Coli Nissle 1917 for the Delivery of Matrix-Tethered Therapeutic Domains to the Gut.” Nature Communications, vol. 10, no. 1, Dec. 2019, https://doi.org/10.1038/s41467-019-13336-6. Accessed 17 Apr. 2021.
- Tang, Tzu-Chieh, et al. “Materials Design by Synthetic Biology.” Nature Reviews Materials, vol. 6, no. 4, 23 Dec. 2020, pp. 332–350, https://doi.org/10.1038/s41578-020-00265-w.
- Tumas, Sarunas, et al. “Engineered E. Coli Nissle 1917 for Delivery of Bioactive IL-2 for Cancer Immunotherapy.” Scientific Reports, vol. 13, no. 1, 2 Aug. 2023, p. 12506, www.nature.com/articles/s41598-023-39365-2, https://doi.org/10.1038/s41598-023-39365-2. Accessed 19 Sept. 2023.