Focusing on MRSA
Staphylococcus aureus is one of the most notorious and ubiquitous bacterial pathogens, causing an incalculable number of simple skin infections worldwide each year, with hundreds of thousands to millions of more serious invasive infections. Among them, methicillin-resistant Staphylococcus aureus (MRSA) is recognized as a "superbug". MRSA can invade the mammary glands, mucous membranes, serous membranes, skin, and internal organs of humans and other animals (cattle, chickens, horses, dogs, pigs, and cats), causing serious disease and is a focal point of infection control worldwide.
Food poisoning is a special case of acute Staphylococcus aureus infection, which is clinically manifested by gastrointestinal symptoms such as nausea, vomiting, abdominal pain, and diarrhea. Food poisoning caused by Staphylococcus aureus has been reported one after another, with food poisoning caused by Staphylococcus aureus accounting for about 25% of foodborne microbial food poisoning outbreaks.
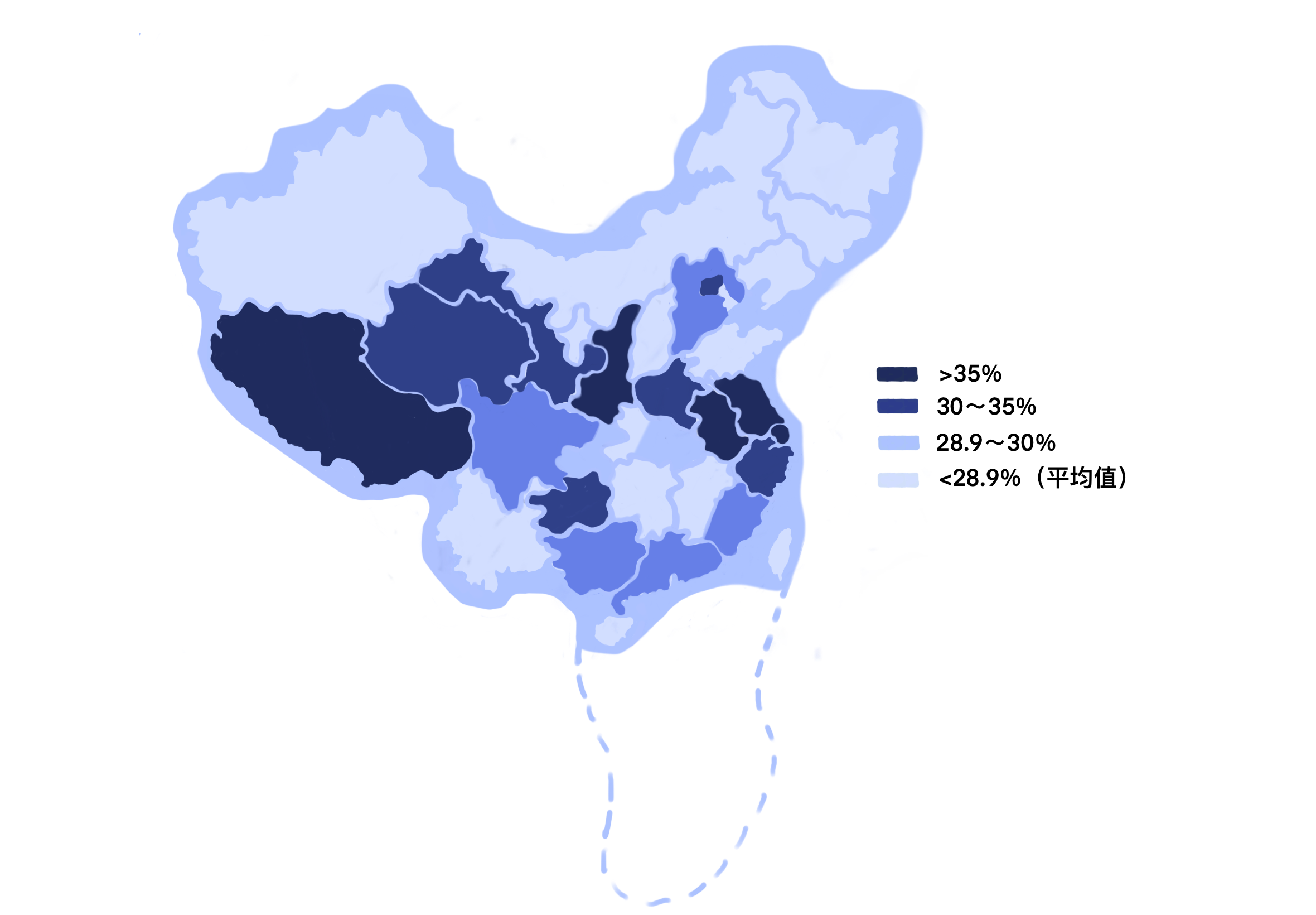
According to the 2022 CARSS report, the national average detection rate of MRSA was 28.9%, a decrease of 0.5 percentage points compared with 2021, and there were some differences between regions, with the highest detection rate in Tibet Autonomous Region (44.0%) and the lowest in Liaoning Province (16.0%) (Figure2)1.
Therefore, we urgently need to find more reasonable treatment and control methods. Synthetic biology offers a promising approach to this problem.
Current Situation
Due to drug resistance, there are many limitations in the treatment of MRSA, and most of the current treatment methods are antibiotic treatment, such as vancomycin, ipamicin, clindamycin, etc.
However, the price of linezolid and vancomycin is relatively high, and long-term use may also cause bone marrow atrophy. Teicoplanin otorenal toxicity is lower than vancomycin, but the onset of action is slow, so it cannot be used as a first-line drug for severe infection. At the same time, the emergence of antimicrobial resistance has weakened the effectiveness of existing antibiotics. Prevention and treatment of MRSA infection are therefore crucial. At this time, antimicrobial peptides have been widely studied as a promising antimicrobial strategy.
| therapy type | costs(CNY) | Application mode | side effect | Whether drug resistance can develop | draw when apply to treat MRSA |
|---|---|---|---|---|---|
| Atropine | 1-7/mg | Injection,less convenient | Constipation,reduced sweating, etc | no | MRSA cannot be completely killed |
| Vancomycin | 200/round | Injection,less convenient | Rash, nausea, phlebitis, etc | yes | more expensive, can develop drug resistance |
| Teicoplanin | 500-700/round | intravenous administration | yes | requires loading dose, which will produce resistance | |
| Linezolid | 4000-5000/round | Intravenous or oral | Diarrhea, headaches and nausea | no | expensive |
Why Super Eco.?
Recognizing the inherent limitations of current MRSA treatments, we embarked on a journey to harness the power of synthetic biology to create novel antimicrobial peptide therapies, and we aptly named our product super Eco. Our main goal is to engineer bacteria so that they can efficiently secrete antimicrobial peptides to specifically kill Staphylococcus aureus, and are equipped with a sensing switch and concentration feedback to make it biosafe.
Our motivation to address the shortcomings of existing MRSA treatments led us to explore the enormous potential of synthetic biology. A significant advantage of synthetic biology is its ability to precisely control downstream output signals by combining a myriad of different control elements. This inherent flexibility allows us to build customized control systems within the host organism, like assembling building blocks, for therapeutic purposes.
The name of our product and mascot, super Eco., succinctly sums up the superiority of our project design. Our innovative approach to MRSA treatment is firmly rooted in the principles of synthetic biology, and we have successfully achieved our vision of the 6Cs of MRSA treatment: "Affordable, Accessible, Controllable, Sustainable, Potential, Competitive". The following sections will shed light on how our design achieves these goals.
Design
Chassis Organism
We chose E. coli Nissle 1917 as the chassis organism for our design. Escherichia coli Nissle 1917, often abbreviated as "Nissle 1917", is a non-pathogenic strain of Escherichia coli (E. coli), a bacterium commonly found in the human gut. Unlike certain pathogenic strains of E. coli that can cause disease, Nissle 1917 is considered safe for human consumption and has been extensively studied for its potential health benefits. This probiotic strain has been used to treat a variety of gastrointestinal disorders, including irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD). We can effectively prevent food poisoning caused by MRSA by modifying Escherichia coli and taking it in the form of capsules to colonize the large intestine and respond to kill Staphylococcus aureus when it encounters Staphylococcus aureus.
AMP Design
We designed the AMP sequence including a fusion peptide composed of a specific targeting domain and a broad-spectrum AMP domain 2 + narrow-spectrum antibiotic3, so that it can specifically and efficiently kill Staphylococcus aureus. By reviewing the literature, we designed the corresponding sequences: LLGSLLKLLPKLLCASYFCRWWWLL-NH2 (C14-C19), which ensures that the antimicrobial peptide has excellent salt tolerance and biocompatibility.
AMP Circuit
In order to better achieve the goal of targeted anti-Staphylococcus aureus therapy and reduce antibiotic contamination, an efficient, accurate, sensitive and safe control system is necessary. We've tried at all stages of AMP synthesis. In the induction link, we used the quorum sensing system of Staphylococcus aureus to design a highly specific sensing switch, which is more accurate. In the initiation of expression, we designed a fast-response expression pathway with positive feedback characteristics by simultaneously expressing the CI protein and then relieving its own transcriptional repression through an inverter, which is more sensitive. In the stable expression state, we used the mechanism of self-negative regulation of CI protein to design a negative feedback regulatory pathway that can inhibit the expression level of AMP, which is safer.
P/N Feedback LoopIn order to effectively control the concentration of antimicrobial peptides, reduce the occurrence of drug resistance, and reduce the impact on the original intestinal microbes, we introduced the lysogenic pathway pathway pathway of bacteriophages to achieve positive feedback response and negative feedback regulation of AMP concentration.
Concentration Module
By introducing a key module in the Agr two-component system of Staphylococcus aureus, quorum sensing is used to achieve a sensitive linear response to Staphylococcus aureus concentrations.
Biofilm
Due to the presence of some dormant Staphylococcus aureus cells (also known as persistent cells) that are encapsulated by its biofilm. These cells remain dormant during antimicrobial therapy and become active once treatment is stopped, causing chronic recurrent infections4. The colonization of engineered probiotics and the hydrogel secreted by the probiotics repair the intestinal barrier, weaken the colonization ability of Staphylococcus aureus that was originally colonized in the host and newly entered the body, enhance the killing ability of antimicrobial peptides, block the reproduction of Staphylococcus aureus, and remove dormant Staphylococcus aureus.
Biosafety
We start from different levels of life and design biosafety modules based on different safety purposes, from the transcription of gene pathways to expression vectors, to suicide mechanisms at the individual level, and finally to the quorum sensing mechanism of populations. The abundance of regulatory means ensures that our drug system has no additional burden on the human body, and the use of the only probiotic in E. coli as a carrier greatly reduces the possibility of heterologous colonization and the risk of bacteria escaping to other parts of the body.
| Life level | Primary Objective | Solution |
|---|---|---|
| Gene | Artificially control the transcriptional expression of genes in extreme environmental or agnostic environments | DD/Shield1 system |
| Expression vectors | Avoid the threat of instability posed by the widespread horizontal gene transfer between microorganisms | Nonmetastatic plasmids |
| Individual | Avoid the risk of E. coli planned colonizing the human intestine being excreted outside the body | Extracorporeal thermosensitive suicide pathway |
| Population | Ensure that AMP precisely responds to Staphylococcus aureus synthesis and controls population density | Quorum Sensing System |
We have added an overall control switch, DD/Shield1 System5, upstream of the key promoters of the synthesis sequence. By applying the exogenous small molecule substance Shield1, the expression of downstream pathways can be inhibited, so as to artificially control the transcriptional expression of genes in extreme environments.
Selection of Plasmids
We looked for non-metastatic plasmids lacking oriT regions according to oriTfinder6, thereby avoiding horizontal gene transfer.
Temperature Suicidal Circuit
We express the thermoreceptor within E. coli and link it to downstream genes that induce cell lysis. Our temperature-sensing suicide has the following characteristics, which is achieved by adding a biphasic steady state switch7.
(1) Silence is maintained during in vitro preparation, transportation, and storage;
(2) After E. coli enters the human body to restore biological activity, the pathway needs to open the perception of temperature.
Quorum Sensing System
The quorum sensing (QS) pathway we designed will express AHL and its receptors, and when the concentration is greater than a certain threshold, it will bind to the regulatory sequence to open the downstream suicide pathway to control the number of cells. In addition, we have also modified the composition of the control sequence to set the activation threshold and prevent QS system leakage, and other optimization measures8.
Conclusion
We thus constructed an engineered Escherichia coli Nissle 1917 that can colonize the intestine, which can specifically recognize Staphylococcus aureus in intestinal infection and kill Staphylococcus aureus by secreting antimicrobial peptides. Our projects have the advantages of 6C: cheap, convenient, controllable, sustainable, potential, and competitive.
Future Aspects
In the future, we will consider genome integration to ensure long-term validity and construct an orthogonal codon system9 to ensure that its genes are not transferred to other microorganisms9.
We will develop transpeptidase-mediated C-terminal PEGylation of intracellular antimicrobial peptides, which can achieve specific C-terminal PEGylation in host cells by biosynthesis of SortaseA-containing PEG compounds. This will further improve the stability of antimicrobial peptides without the development of drug resistance.
Also, we will further weaken or even eliminate adverse reactions by adding modules that neutralize enterotoxins. On the other hand, we are also extending it to the livestock industry to further increase its application.
Reference
- 2022年全国细菌耐药监测报告(简要版). https://www.carss.cn/Report/Details/917.
- Shang, L.; Li, J.; Song, C.; Nina, Z.; Li, Q.; Chou, S.; Wang, Z.; Shan, A., Hybrid Antimicrobial Peptide Targeting Staphylococcus aureus and Displaying Anti-infective Activity in a Murine Model. 2020, 11.
- Mishra, B.; Wang, G. J. J. o. t. A. C. S., Ab initio design of potent anti-MRSA peptides based on database filtering technology. 2012, 134 (30), 12426-9.
- Idrees, M.; Sawant, S.; Karodia, N.; Rahman, A., Staphylococcus aureus Biofilm: Morphology, Genetics, Pathogenesis and Treatment Strategies. 2021, 18 (14), 7602.
- Haugwitz, M.; Garachtchenko, T.; Nourzaie, O.; Gandlur, S.; Sagawa, H., Rapid, on-demand protein stabilization and destabilization using the ProteoTuner™ systems. Nature Methods 2008, 5 (10), iii-iv.
- Li, X.; Xie, Y.; Liu, M.; Tai, C.; Sun, J.; Deng, Z.; Ou, H.-Y., oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res 2018, 46 (W1), W229-W234.
- Elowitz, M. B.; Leibler, S., A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403 (6767), 335-338.
- Ge, C.; Yu, Z.; Sheng, H.; Shen, X.; Sun, X.; Zhang, Y.; Yan, Y.; Wang, J.; Yuan, Q., Redesigning regulatory components of quorum-sensing system for diverse metabolic control. Nat Commun 2022, 13 (1), 2182.
- Zürcher, J. F.; Robertson, W. E.; Kappes, T.; Petris, G.; Elliott, T. S.; Salmond, G. P. C.; Chin, J. W. J. S., Refactored genetic codes enable bidirectional genetic isolation. 2022, 378, 516 - 523.